Handling Imbalanced Data in Machine Learning: Techniques, Examples, and Best Practices-Supervised learning with scikit-learn
Supervised learning with scikit-learn (Part 8)
📚Chapter:3-Data Preprocessing and Pipelines
If you want to read more articles about Supervise Learning with Sklearn, don’t forget to stay tuned :) click here.
Table Content
Definition
Real-life Examples of imbalanced data problem
Why Imbalanced dataset is bad?
Check Imbalanced Problems in Data
Handle Imbalanced dataset problem
Performance metrics
Suitable ML algorithm for imbalanced Problem
Section1- Definition
Defi 1: Classification is one of the most common machine-learning problems. One of the common issues found in datasets that are used for classification is the imbalanced classes issue. Generally, we expect the labels to be relatively even distributed. For classification tasks, one may encounter situations where the target class label is unequally distributed. Such conditions are termed imbalanced data. In reality, the samples we can get may be unsatisfactory.
Def 2: An imbalance occurs when one or more classes have very low proportions/probability in the training data as compared to the other classes. For classification tasks, one may encounter situations where the target class label is unequally distributed.
Def 3: In the context of binary classification, the less frequently occurring class is called the minority class, and the more frequently occurring class is called the majority class [3]. Such conditions are termed imbalanced data. . A data scientist might face some challenges while modeling an imbalanced class dataset. There are various hacks or techniques to handle an imbalanced class data sample prior to modeling including[3].
Def: An imbalanced dataset is when you have, for example, a classification test and 90% of the data is in one class. That leads to problems: an accuracy of 90% can be skewed if you have no predictive power on the other category of data! Here are a few tactics to get over the hump:
1- Collect more data to even the imbalances in the dataset.
2- Resample the dataset to correct for imbalances.
3- Try a different algorithm altogether on your dataset.
Section 2- Real-life Examples of Imbalanced Data Problem
1- Imagine you are trying to build a classification model, and you have two classes: Cats and Dogs. Unfortunately, your data is very skewed: there are 950 cat pictures and 50 dog pictures.[2].If your model classifies every picture as a Cat (dumb, right?), you’ll be 95% accurate! Think about that for a second. The distribution in your dataset becomes a big problem really quickly [2].
2- Suppose we have a diabetes dataset and we have to predict whether the patient has diabetes or not. In this case, our result will either yes or no. As it is a classification machine learning problem, we will see two classes “Diabetes” and “Not Diabetes” in our target or dependent feature of the dataset. Assume we have a dataset of 1000 patients, out of which 900 patients are healthy, do not have diabetes, and 100 patients have diabetes. Here clearly see that we have a majority class i.e patients which don’t have diabetes having 9 times bigger than the minority class i.e diabetes class. Such a type of dataset where one class has much more majority than the other class is called an Imbalanced dataset [5]
Section 3- Why Imbalanced dataset is bad?
So why does imbalance lead to poor model performance? Because any algorithm cannot obtain enough information from a class with a small sample size to make accurate predictions.
The uneven distribution of the corresponding variables reduces the accuracy of the algorithm, and the prediction accuracy for small classes will be very low.
The algorithm itself is error-driven, that is, the goal of the model is to minimize the overall error, and the contribution of small classes to the overall error is very low.
The algorithms themselves assume that the class distribution of the data set is balanced, and they may also assume that different classes of errors bring the same loss.
Our model gives us poor predictive performance, specifically for the minority class if we don’t handle the imbalanced data.[1].
Due to the presence of class imbalance the model might become biased towards the majority class data sample
When we have an imbalanced dataset and if we build a machine learning model over imbalanced data, then there is a high chance of getting misclassification i.e incorrect classification[5].
Take a binary classification problem, for example, we might encounter too few positive samples in the training dataset, called minority class. And most machine learning techniques will ignore, and in turn have poor performance on, the minority class, although typically it is the performance on the minority class that is most important.
Section 4-Check Imbalanced Problems in Data
import matplotlib.pyplot as plt
fig = plt.figure(figsize = (8,5))
Train_data.RESULT_TEXT.value_counts(normalize = True).plot(kind='bar', color= ['skyblue','navy'], alpha = 0.9, rot=0)
plt.title('Results Status Negative(0) and Positive(1) in the Imbalanced Dataset')
plt.show()Section 5- Handle Imbalanced dataset Problem
There are the following types to handle the imbalanced problem.
5. 1- Resample the dataset
Def: Resampling is one of the most popular techniques to handle an imbalanced dataset. The main objective of resampling is to balance the dataset either by increasing the frequency of the minority class or by decreasing the frequency of the majority class [5].
Disadvantages
Keep in mind that over and undersampling introduce a bias into your dataset: You are changing the data distribution by arbitrarily messing with the existing samples. This might have consequences, so think about this carefully [2].
5.1.1- Oversampling
Def: Sampling is used to increase the minority class by randomly duplicating them. The idea is to duplicate some random examples from the minority class thus this technique does not add any new information from the data.[13] Over Sampling is used to increase the minority class by randomly duplicating them. Upsampling or Oversampling refers to creating artificial minority class data points to balance the distribution between the majority and minority class samples.
Background: For example: Take the pictures we have multiple times [2]. This method mainly deals with small classes. Use repeated observations to balance the data. On the contrary, oversampling is used when the quantity of data is insufficient. It tries to balance dataset by increasing the size of rare samples. Rather than getting rid of abundant samples, new rare samples are generated by using e.g. repetition, bootstrapping or SMOTE (Synthetic Minority Over-Sampling Technique) [1].It is noteworthy that cross-validation should be applied properly while using the over-sampling method to address imbalance problems. Keep in mind that over-sampling takes observed rare samples and applies bootstrapping to generate new random data based on a distribution function. If cross-validation is applied after over-sampling, basically what we are doing is overfitting our model to a specific artificial bootstrapping result. That is why cross-validation should always be done before over-sampling the data, just as how feature selection should be implemented. Only by resampling the data repeatedly, randomness can be introduced into the dataset to make sure that there won’t be an overfitting problem [7]
Advantages
There will not be any information loss using this method,
Disadvantages
but the addition of small-type repeated samples will easily lead to over-fitting, and the calculation time and storage overhead will also increase[4].
The main disadvantage of oversampling is, it will cause overfitting the machine learning model as we are duplicating the records of minority classes randomly [5].
Oversampling techniques
There are various oversampling techniques that can be used to generate artificial or duplicate minority class data points.
Random Oversample
SMOTE
ADASYN
A-RandomOverSampler
Importing library RandomOverSampler for Over Sampling and then fitting independent features X and dependent feature y.
from imblearn.over_sampling import RandomOverSampler
ros = RandomOverSampler()
X_os, y_os = ros.fit_resample(X,y)[10]
from sklearn.datasets import make_classification
X, y = make_classification(n_samples=5000, n_features=2, n_informative=2,
n_redundant=0, n_repeated=0, n_classes=3,
n_clusters_per_class=1,
weights=[0.01, 0.05, 0.94],
class_sep=0.8, random_state=0)
from imblearn.over_sampling import RandomOverSampler
ros = RandomOverSampler(random_state=0)
X_resampled, y_resampled = ros.fit_resample(X, y)
from collections import Counter
print(sorted(Counter(y_resampled).items()))
[(0, 4674), (1, 4674), (2, 4674)]B- SMOTE
Synthetic Minority Oversampling Technique (SMOTE) is one of the most popular oversampling techniques that is developed by Chawla et al. (2002). SMOTE addresses class imbalance by generating synthetic samples for the minority class. Addressing Class Imbalance: SMOTE tackles the problem where one class (usually minority) has far fewer samples than another class (majority), which can lead to biased models.
Unlike random oversampling which only duplicates some random examples from the minority class, SMOTE generates examples based on the distance of each data (usually using Euclidean distance) and the minority class nearest neighbors, so the generated examples are different from the original minority class. This method is effective because the synthetic data that are generated are relatively close with the feature space on the minority class, thus adding new “information” on the data, unlike the original oversampling method [13]. It is an oversampling technique where the samples are generated for the minority class. These are not duplicates or replicates of existing minority class data. It creates new points for the minority class [23]. We can perform oversampling of the minority class using SMOTE technique and further perform undersample using Edited Nearest Neighbour (ENN). The advantage of performing the undersampling technique — ENN after oversampling is that it cleans or removes the noise majority of class data points [4].A part from the random sampling with replacement, there are two popular methods to over-sample minority classes: (i) the Synthetic Minority Oversampling Technique (SMOTE) [CBHK02] and (ii) the Adaptive Synthetic (ADASYN) [HBGL08] sampling method. These algorithms can be used in the same manner:
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state=42)
X_res, y_res = sm.fit_resample(X_train, y_train)import matplotlib.pyplot as plt
fig = plt.figure(figsize = (8,5))
Train_data.RESULT_TEXT.value_counts(normalize = True).plot(kind='bar', color= ['skyblue','navy'], alpha = 0.9, rot=0)
plt.title('Results Status Negative(0) and Positive(1) in the Imbalanced Dataset')
plt.show()from imblearn.over_sampling import SMOTENC
smote_nc = SMOTENC(categorical_features=[0, 2], random_state=0)
X_resampled, y_resampled = smote_nc.fit_resample(X, y)
print(sorted(Counter(y_resampled).items()))
#[(0, 30), (1, 30)]
print(X_resampled[-5:])
Output
[['A' 0.5246469549655818 2]
['B' -0.3657680728116921 2]
['B' 0.9344237230779993 2]
['B' 0.3710891618824609 2]
['B' 0.3327240726719727 2]]from imblearn.over_sampling import SMOTE, ADASYN
X_resampled, y_resampled = SMOTE().fit_resample(X, y)
print(sorted(Counter(y_resampled).items()))
[(0, 4674), (1, 4674), (2, 4674)]
clf_smote = LinearSVC().fit(X_resampled, y_resampled)
X_resampled, y_resampled = ADASYN().fit_resample(X, y)
print(sorted(Counter(y_resampled).items()))
[(0, 4673), (1, 4662), (2, 4674)]
clf_adasyn = LinearSVC().fit(X_resampled, y_resampled)C- ADASYN
ADASYN (Adaptive Synthetic Minority Oversampling Technique) is an oversampling algorithm used to address imbalanced data problems in machine learning. It focuses on generating synthetic minority class data points in areas with higher decision boundaries, aiming to improve the model’s performance on the minority class.
Benefits of ADASYN:
Improves model performance on the minority class.
Reduces bias caused by imbalanced data.
Adaptively focuses on areas with higher decision boundaries.
Limitations of ADASYN:
May overfit the minority class.
Can be computationally expensive for large datasets.
Here’s how to implement ADASYN in Python:
#1. Install libraries:
!pip install imbalanced-learn
#2. Import libraries and load data:
import pandas as pd
from imblearn.over_sampling import ADASYN
# Load your imbalanced data
data = pd.read_csv("your_data.csv")
X = data.drop("target", axis=1)
y = data["target"]
# 3. Instantiate ADASYN object:
# Specify desired ratio of minority class over majority class after resampling
ratio = "auto" # equalize all classes
# Instantiate ADASYN object
adasyn = ADASYN(sampling_strategy=ratio)
# 4. Oversample data:
# Resample X and y data
X_res, y_res = adasyn.fit_resample(X, y)
D) Borderline-SMOTE
Because we want to provide our model with as much data as possible, we will use a technique called Borderline-SMOTE. Borderline-SMOTE(Borderline Synthetic Minority Oversampling Technique). The reason that Borderline-SMOTE was used as opposed to SMOTE is that it takes synthetically generates data in between different elements in the minority class, while ignoring outliers. Outliers can be defined as features which neighbour more majority points, than minority ones.
from imblearn.over_sampling import BorderlineSMOTE
X_train_Before= X_train
y_train_Before= y_train
borderlineSMOTE = BorderlineSMOTE(k_neighbors = 10, random_state = 42)
X_train, y_train = borderlineSMOTE.fit_resample(X_train_Before,y_train_Before)5.1.2- Undersampling
Def: the under-sampling technique was developed in 1976 by Tomek [13]. It is conducted by removing some random examples from the majority class, at cost of some information in the original data being removed as well[13].Under Sampling is used to decrease the majority class by randomly eliminating the majority class. This is done until majority and minority class instances are balanced out [5]. Down-sampling or undersampling refers to the removal of majority class data points to balance the target class distribution. for example, Take some of the pictures we have, and discard the rest [2]
Background: It is one of a modification from Condensed Nearest Neighbors (CNN). Talking about the Condensed Nearest Neighbor Rule, examples are selected randomly, especially initially, which results in the retention of unnecessary samples. It is based on the nearest-neighbor (NN) rule [13].
Benefits of Undersampling:
This method is used when the quantity of data is sufficient.By keeping all samples in the rare class and randomly selecting an equal number of samples in the abundant class, a balanced new dataset can be retrieved for further modelling [7]
This method is mainly to deal with large categories. Use to reduce the number of observations in large categories to balance the data set. It is suitable when the overall data set is large,
This method can also reduce the calculation time and storage overhead (the training set samples are less).
Disadvantages of Undersampling:
The main disadvantage of under-sampling is that we lose the information of data. We can use under-sampling when we have millions of records [5].
Under Sampling is used to decrease the majority class by randomly eliminating the majority class.
This is done until majority and minority class instances are balanced out [5].The problem that arises with imbalanced datasets is that they are predominately composed of majority class (normal examples) with only a small percentage of minority class (abnormal or interesting examples). This makes the model unable to learn from minority class well. Sometimes, this minority class can contain vital information like disease detection dataset, churn dataset, and fraud detection dataset[13].
There are various Undersampling techniques
Random Undersampling
NearMiss
Tomek Links
Cluster-Centroid Undersampling
Edited Nearest Neighbors
A- Random Undersampling
This is the simplest method. It randomly removes samples from the majority class until the desired class balance is achieved.
from sklearn.utils.resample import RandomUnderSampler
# Define the undersampler
rus = RandomUnderSampler(sampling_strategy='majority')
# Undersample the data
X_resampled, y_resampled = rus.fit_resample(X, y)B- NearMiss
Importing library NearMiss for Under Sampling and then fitting independent features X and dependent feature y [5].This method selects examples from the majority class that are closest to the minority class samples, based on a specific distance metric. This helps to retain informative data from the majority class while achieving balance.
from imblearn.under_sampling import NearMiss
nm = NearMiss()
X_us, y_us = nm.fit_resample(X,y)from imblearn.under_sampling import NearMiss
# Define the undersampler
nmiss = NearMiss(sampling_strategy='majority')
# Undersample the data
X_resampled, y_resampled = nmiss.fit_resample(X, y)C. Tomek Links
This method identifies and removes pairs of majority class samples that are nearest neighbors to each other, while not being nearest neighbors to any minority class samples.
from imblearn.under_sampling import TomekLinks
# Define the undersampler
tl = TomekLinks(sampling_strategy='majority')
# Undersample the data
X_resampled, y_resampled = tl.fit_resample(X, y)D. Cluster-Centroid Undersampling:
This method clusters the majority of class data and then removes all samples except the cluster centroids. This helps to retain representative samples from the majority class while reducing its size.
from imblearn.under_sampling import ClusterCentroids
# Define the undersampler
cc = ClusterCentroids(sampling_strategy='majority')
# Undersample the data
X_resampled, y_resampled = cc.fit_resample(X, y)E. Edited Nearest Neighbors:
This method removes majority class samples that are closer to minority class samples than any other majority class sample. This helps to remove outliers from the majority class that might negatively impact the model.
from imblearn.under_sampling import EditedNearestNeighbours
# Define the undersampler
enn = EditedNearestNeighbours(sampling_strategy='majority')
# Undersample the data
X_resampled, y_resampled = enn.fit_resample(X, y)Choosing the Right Method:
The best undersampling method for your dataset will depend on various factors, including the size and distribution of the data, the specific classification task, and your computational resources.
Here are some additional points to consider:
Random undersampling is simple and efficient, but it can discard valuable information from the majority class.
NearMiss and Edited Nearest Neighbors are more sophisticated methods that can retain more information, but they can also be more computationally expensive.
Cluster-Centroid Undersampling can be effective for datasets with well-separated clusters, but it might not be suitable for datasets with overlapping clusters.
Tomek Links can be effective for removing redundant data from the majority class, but it can be sensitive to outliers.
It is recommended to experiment with different undersampling methods and compare their performance on your specific dataset to choose the most effective one.
5.2- Combine both techniques
It is also possible to combine the two methods of oversampling and under-sampling. The large class uses und-ersampling without replacement, and the small class uses oversampling with replacement.
5.2.1 Edited Nearest Neighbour (ENN)
The advantage of performing the undersampling technique — ENN after oversampling is that it cleans or removes the noise majority class data points [4]
smt = SMOTEENN(random_state=42)
X_res, y_res = smt.fit_resample(X_train, y_train)5.2.2 — SMOTE-Tomek Links
Similarly, we can perform oversampling of the minority class using SMOTE technique and further undersample or perform cleaning using the Tomek Links technique [4].
smt = SMOTETomek(random_state=42)
X_res, y_res = smt.fit_resample(X_train, y_train)A combination of over-sampling the minority (abnormal) class and under-sampling the majority (normal) class can achieve better classifier performance than only under-sampling the majority class. This method was first introduced by Batista et al. (2003). This method combines the SMOTE ability to generate synthetic data for minority class and Tomek Links ability to remove the data that are identified as Tomek links from the majority class (that is, samples of data from the majority class that is closest with the minority class data). We can say that over-sampling is done using SMOTE while cleaning is done using Tomek links. To understand this method better, let’s take a look at the code[13].
Import libraries.
import pandas as pd
import numpy as np
from imblearn.pipeline
import Pipeline
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_validate
from sklearn.model_selection importRepeatedStratifiedKFold
from sklearn.ensemble import RandomForestClassifier
from imblearn.combine import SMOTETomek
from imblearn.under_sampling import TomekLinksNow, generate synthetic data
#Dummy dataset study case
X, Y = make_classification(n_samples=10000, n_features=4, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1)Normally, a model will fail to learn the minority class but by using the SMOTE-Tomek Links method, we can improve the model’s performance to handle imbalanced data.
## With SMOTE-Tomek Links method
# Define model
model=RandomForestClassifier(criterion='entropy')
# Define SMOTE-Tomek Links resample=SMOTETomek(tomek=TomekLinks(sampling_strategy='majority'))
# Define pipeline
pipeline=Pipeline(steps=[('r', resample), ('m', model)])
# Define evaluation procedure (here we use Repeated Stratified K-Fold CV)
cv=RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
# Evaluate model
scoring=['accuracy','precision_macro','recall_macro'] scores = cross_validate(pipeline, X, Y, scoring=scoring, cv=cv, n_jobs=-1)
# summarize performance
print('Mean Accuracy: %.4f' % np.mean(scores['test_accuracy'])) print('Mean Precision: %.4f' % np.mean(scores['test_precision_macro'])) print('Mean Recall: %.4f' % np.mean(scores['test_recall_macro']))The result is as follows:
Mean Accuracy: 0.9805
Mean Precision: 0.6499
Mean Recall: 0.8433The accuracy and precision metrics might decrease, but we can see that the recall metric is higher, it means that the model performs better to correctly predict the minority class label by using SMOTE-Tomek Links to handle the imbalanced data.
I have computed the CV AUC-ROC, Test AUC-ROC, Test Precision, and Test Recall metrics for a sample dataset before and after performing SMOTE, SMOTETomek, and SMOTEENN respectively.
Further recorded the metrics numbers for a Decision Tree Classifier.[4]
Oversample one class while we also undersample the other [2].
5.3 Collect more data
In our example, we might be able to go and find more dog pictures to add to the dataset and cut down the difference. Unfortunately, collecting more data is not always an option. But sometimes it is, and yet many teams resort to synthetic augmentation prematurely.[2].
5.4- Augment the dataset with synthetic data
Only when collecting more data is not feasible, do I look into making up fake samples. This is a good approach when working with unstructured data (think images, videos, text, audio).[2] For example, you could create new dog pictures by transforming the ones you already have:
Changing the contrast of the image
Doing a horizontal flip
Rotating the picture slightly both ways
Adding some noise
Combining these techniques could generate many new samples that look as realistic as the original pictures. For structured data (think tabular data,) augmentation is way harder and sometimes impossible [2]
5.5- Adjusting the weight of each class appropriately you
could modify your model’s loss function to weigh each class differently to account for the imbalances. Check the sample_weight attribute on Kera’s fit() function for an example [2].Cost-Sensitive Learning is another related technique that you could also use to penalize the result of your model depending on the weight of each class [2]
5.6- Ensemble Different Resampled Datasets
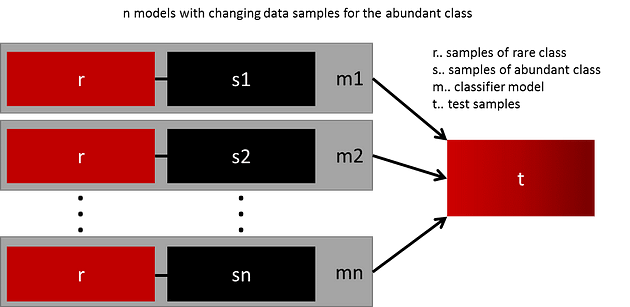
The easiest way to successfully generalize a model is by using more data. The problem is that out-of-the-box classifiers like logistic regression or random forest tend to generalize by discarding the rare class. One easy best practice is building n models that use all the samples of the rare class and n-differing samples of the abundant class. Given that you want to ensemble 10 models, you would keep e.g. the 1.000 cases of the rare class and randomly sample 10.000 cases of the abundant class. Then you just split the 10.000 cases in 10 chunks and train 10 different models [7].
This approach is simple and perfectly horizontally scalable if you have a lot of data since you can just train and run your models on different cluster nodes. Ensemble models also tend to generalize better, which makes this approach easy to handle.
5.7-Resample with Different Ratios
The previous approach can be fine-tuned by playing with the ratio between the rare and the abundant class. The best ratio heavily depends on the data and the models that are used. But instead of training all models with the same ratio in the ensemble, it is worth trying to ensemble different ratios. So if 10 models are trained, it might make sense to have a model that has a ratio of 1:1 (rare: abundant) and another one with 1:3, or even 2:1. Depending on the model used this can influence the weight that one class gets [7].
5.8-Cluster the abundant class
An elegant approach was proposed by Sergey on Quora. Instead of relying on random samples to cover the variety of the training samples, he suggests clustering the abundant class in r groups, with r being the number of cases in r. For each group, only the medoid (center of cluster) is kept. The model is then trained with the rare class and the medoids only [7].
6-Performance metrics
Applying inappropriate evaluation metrics for models generated using imbalanced data can be dangerous.
Accuracy:
Accuracy is not a good performance metric when you have an imbalanced dataset [2]. Imagine our training data is the one illustrated in graph above. If accuracy is used to measure the goodness of a model, a model which classifies all testing samples into “0” will have an excellent accuracy (99.8%), but obviously, this model won’t provide any valuable information for us.
Instead, and depending on your specific problem, you should look at any of the following:
Precision, Recall, F-Score, Confusion Matrix, ROC Curves, MCC , AUC
7-Suitable ML algorithm for imbalanced Problem
Most machine learning models actually get overwhelmed by the majority class, as it expects the classes to be somewhat balanced. It’s like asking a student to learn both algebra and trigonometry equally well but giving him only 5 solved problems of trigonometry to learn from compared to 1000 solved problems in algebra. The patterns of the minority class get buried. This literally becomes the problem of finding a needle from the haystack [3]
Decision Tree-based models are excellent at handling imbalanced classes. When dealing with structured data, that might be all you need [2].
Please Follow and 👏 Subscribe for the story courses teach to see latest updates on this story
If you want to learn more about these topics: Python, Machine Learning Data Science, Statistic For Machine learning, Linear Algebra for Machine learning Computer Vision and Research
Then Login and Enroll in Coursesteach to get fantastic content in the data field.
Stay tuned for our upcoming articles where we will explore specific topics related to Machine learning with scikit-learn in more detail!
Remember, learning is a continuous process. So keep learning and keep creating and Sharing with others!💻✌️
Note:if you are a Supervised learning with scikit-learn’ export and have some good suggestions to improve this blog to share, you write comments and contribute.
if you need more update about Supervised learning with scikit-learn’ and want to contribute then following and enroll in following
👉Course: Supervised learning with scikit-learn
Source
2–1_28_2020_Supervised_learning_with_Sklearn.ipynb
3-Data_Processing_in_Python_.ipynb
4- Python Tricks for Data Science
6- Approaches to Data Imputation(Unread)
8-Scikit-Learn (Python): 6 Useful Tricks for Data Scientists